Biochemistry
Join our mailing listWhere to start?
Start by enrolling in our universal onramp course Foundations in Python & Machine Learning, followed by Deep Learning I. Depending on the research topic you choose you will take courses introducing your topic of interest
All of our programs START HERE:
- Enroll in our "Onramp course": Foundations in Python & Machine Learning
- You can start at any age between 5th and 11th grade. Course cohorts will be grouped by age
Choose a research topic
You can choose from specific areas of interest within the broad field of Biochemistry.
-
Computational protein structure, folding, and design
-
Enzyme kinetics & catalytic mechanism modeling
-
Drug discovery & mol. docking
-
Structure-based drug design
-
Metabolic pathway modeling
-
Protein–ligand interaction
Need more help enrolling?
Schedule a 30 minute consultation.
We will walk you through our program structure and program philosophy to help you choose a track for your student.
We'll answer all your questions including whether our programs are right for your child.
No prior research experience required, however these courses assume deep intellectual curiosity, sustained effort, and a willingness to engage with very challenging material.
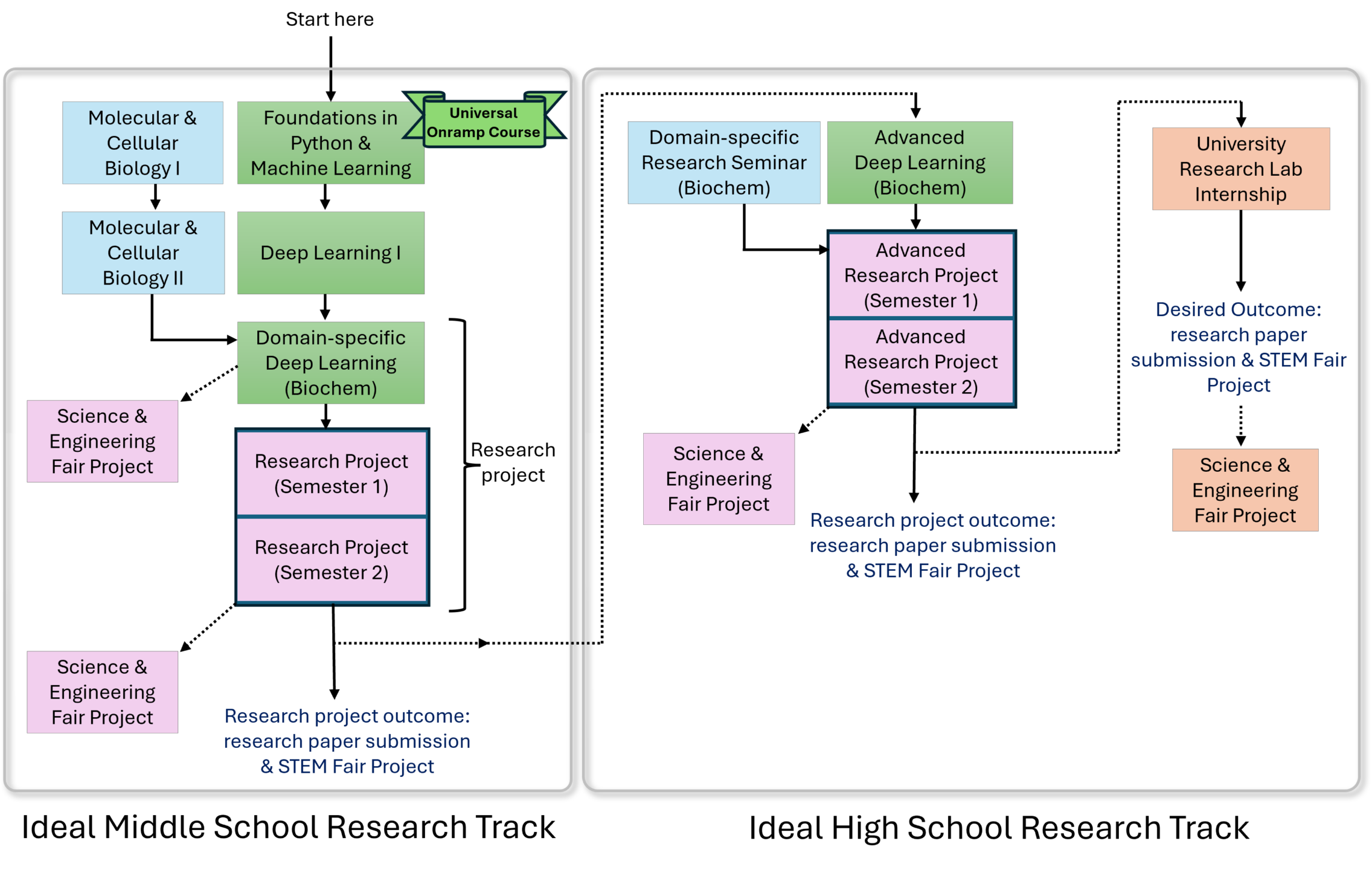
Our comprehensive yet flexible research tracks
Our flexible curricula start with foundational courses in programming & machine learning and domain-specific course work, followed by research programs designed to lead to research paper publication and science fair competition project entries.
Start at any age between 5th and 11th grade. Course cohorts will be grouped by age
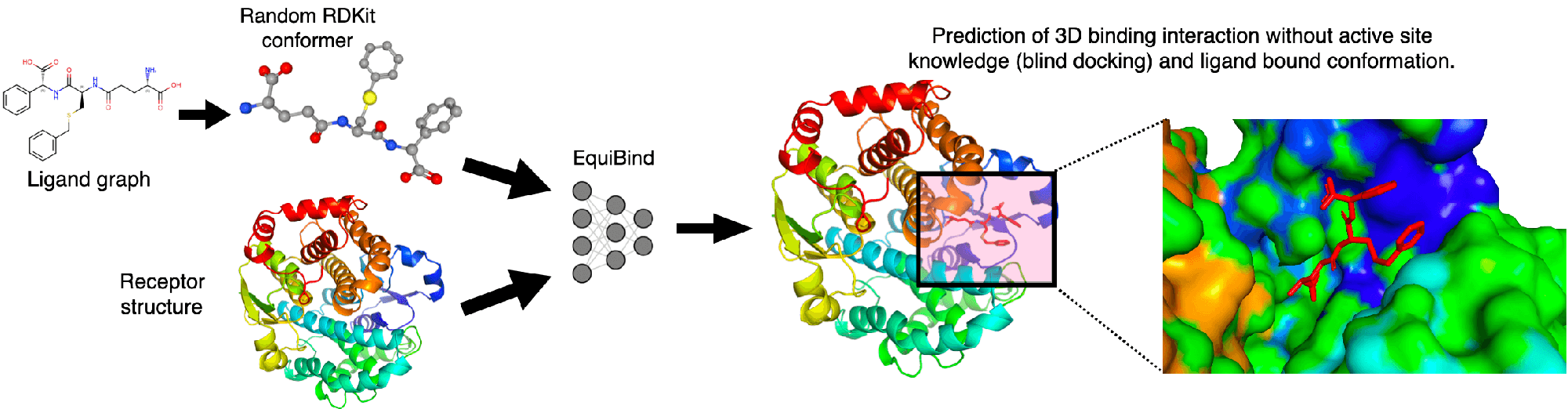
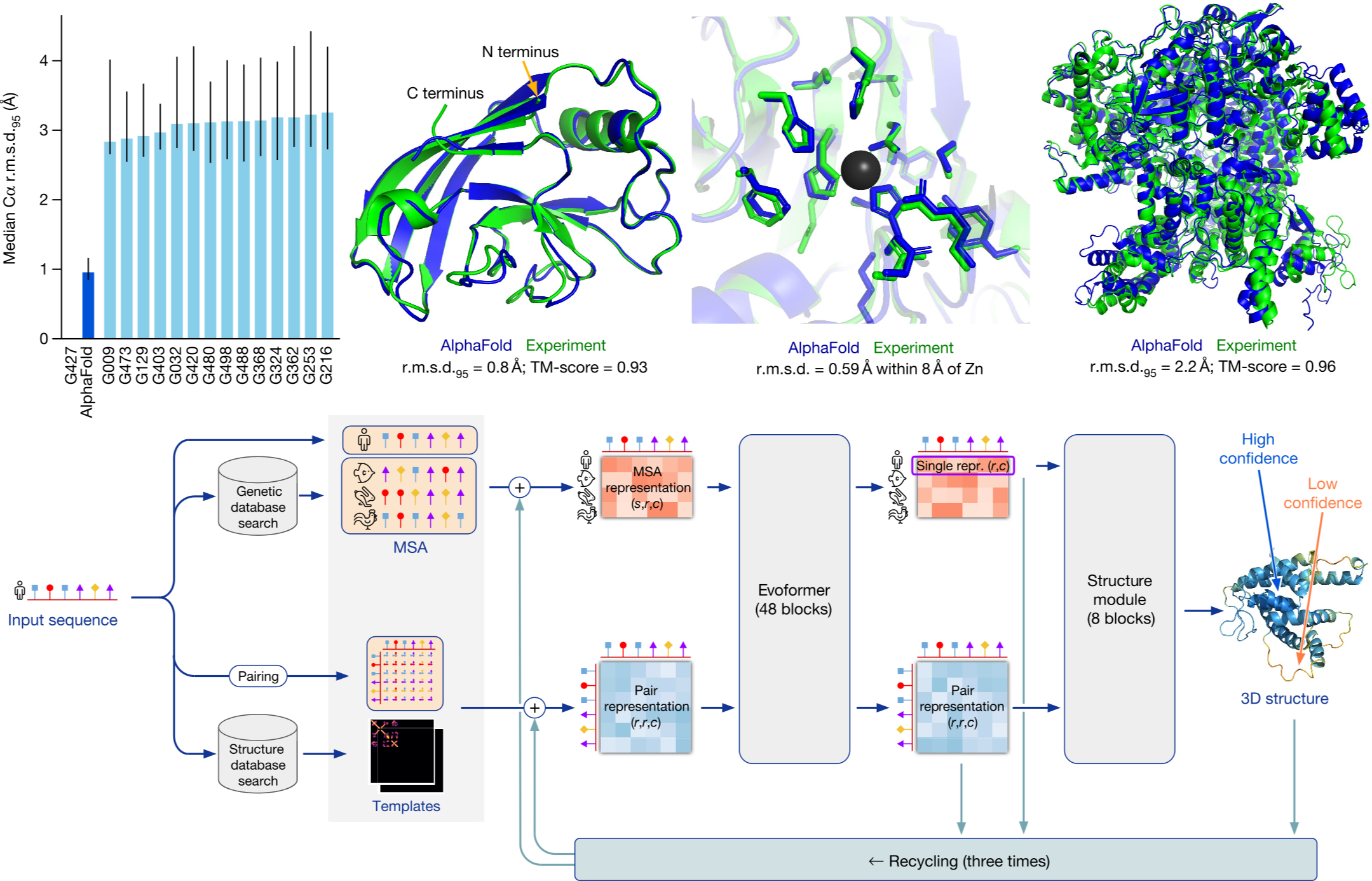
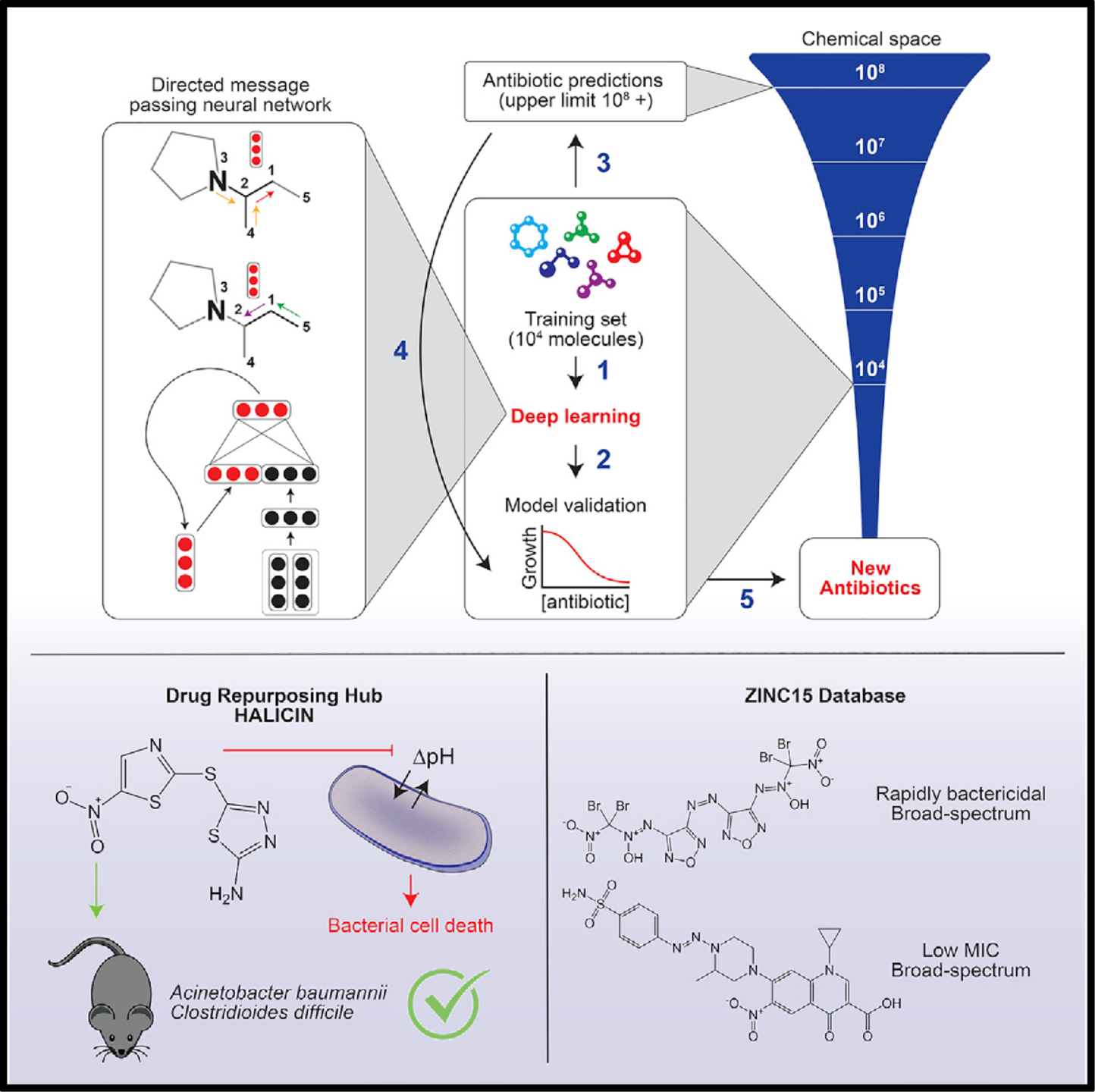
State-of-the-art biochemistry research (below) your student can learn
References for the figures above in order of their appearance. These are state-of-the-art research papers in biochemistry and top high school students have understood research at this level and engaged in research using similar techniques (typically with university mentors)
[1] Stärk, Hannes, et al. "Equibind: Geometric deep learning for drug binding structure prediction." International conference on machine learning. PMLR, 2022.
[2] Jumper, John, et al. "Highly accurate protein structure prediction with AlphaFold." nature 596.7873 (2021): 583-589.
[3] Stokes, Jonathan M., et al. "A deep learning approach to antibiotic discovery." Cell 180.4 (2020): 688-702.
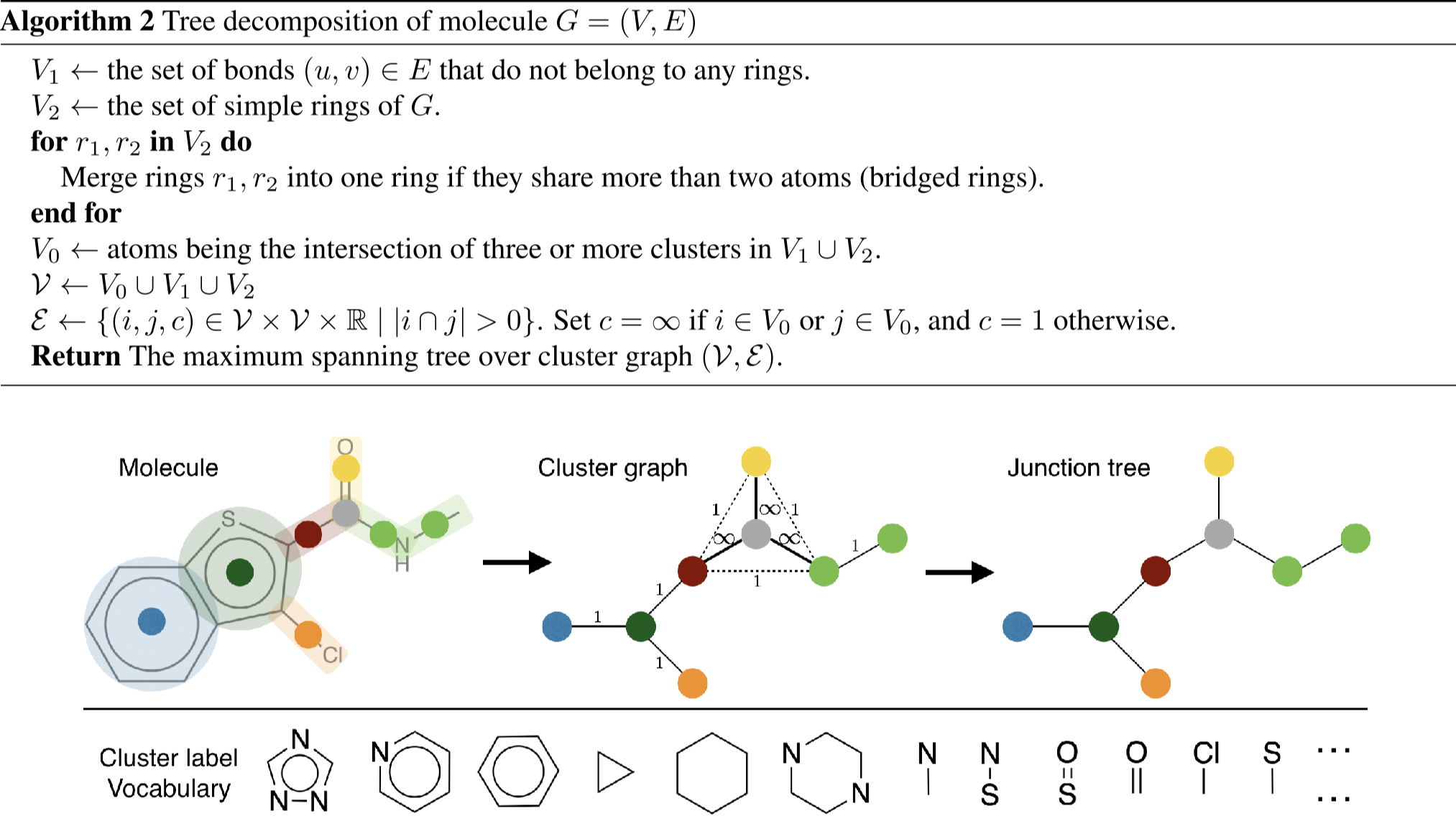
[4] Jin, Wengong, Regina Barzilay, and Tommi Jaakkola. "Junction tree variational autoencoder for molecular graph generation." International conference on machine learning. PMLR, 2018.
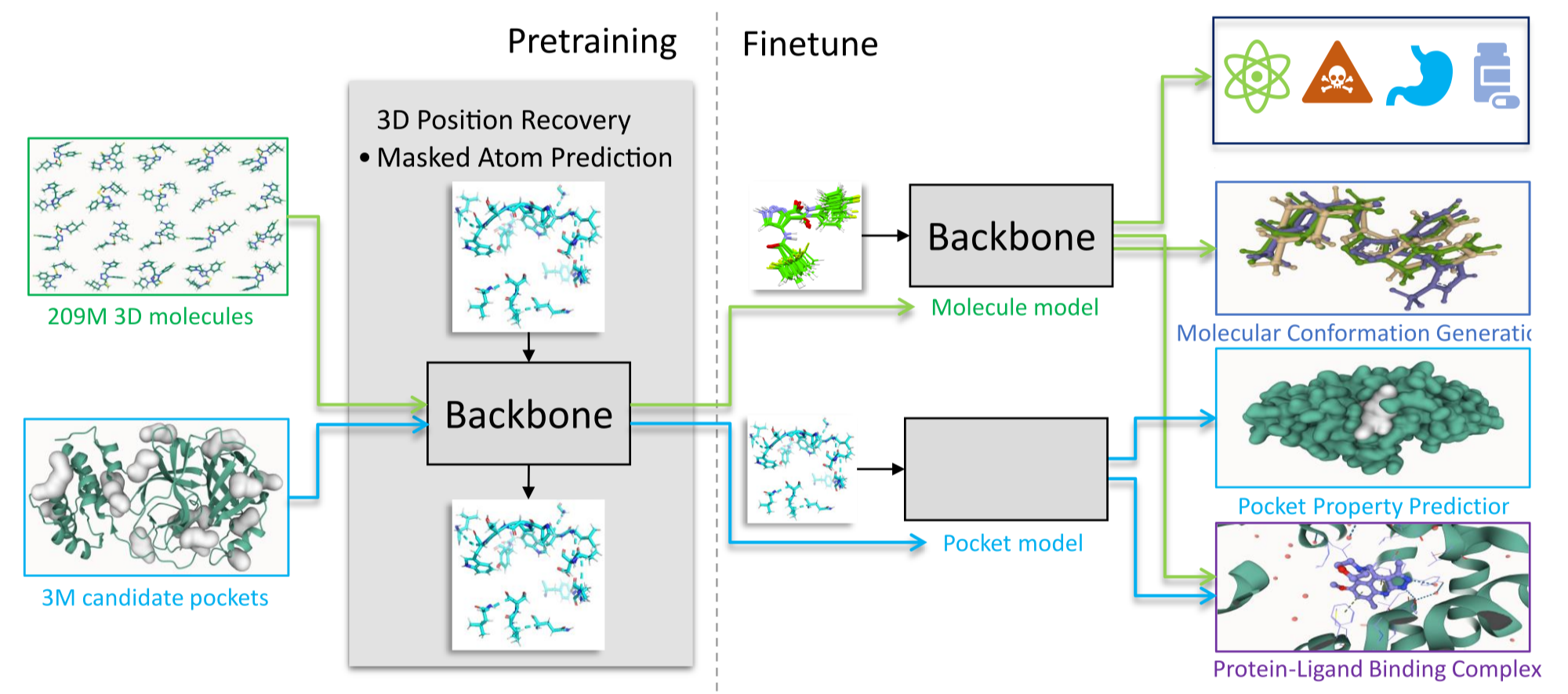
[5] Zhou, Gengmo, et al. "Uni-mol: A universal 3d molecular representation learning framework." The eleventh international conference on learning representations. 2023.